-
Aiding the design of next-generation RAF inhibitors
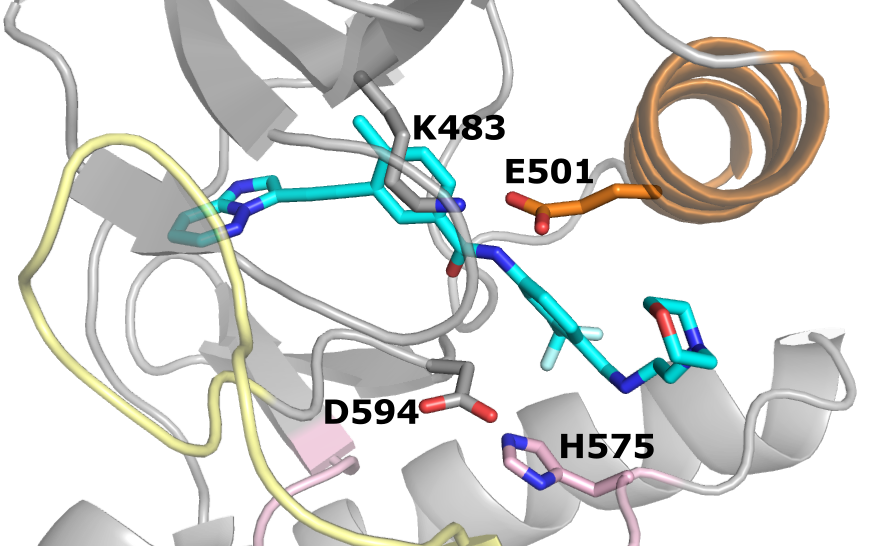
Clayton et al, eLife, 2024
We discovered how dimer selectivity of BRAF inhibitors is linked to the remarkable allostery.

-
ML Models to Interrogate Proteomewide Covalent Ligandabilities
Ruibin Liu et al, JACS Au, 2024
We present a new database and ML models for predicting cysteine-ligandable hot spots in proteins.

-
Aarion won the Department of Pharmaceutical Sciences Merit Award!
Passion. Dedication. Hard work. Congratulations, Aarion! What a great start of the 4th year of PhD study!

-
Direct observation of chitin’s self assembly in silico
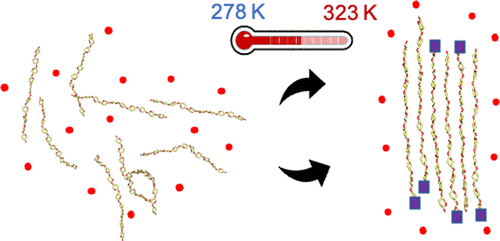
Romany et al, Chem Mater, 2023
De novo self assembly simulations of chitin revealed the mechanism of inverse temperature dependence and polymorphism.

-
Effective quantum descriptors for Michael acceptors
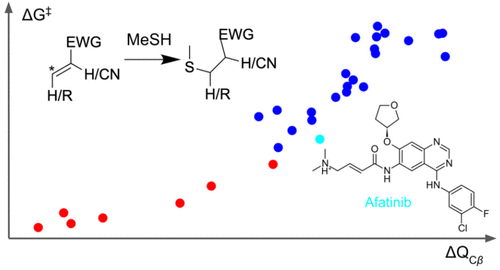
Liu et al, J Chem Inf Model, 2023
Low-cost quantum descriptors can predict the SAR of Michael acceptor warheads.

-
Towards covalently drugging the ERK pathway kinases
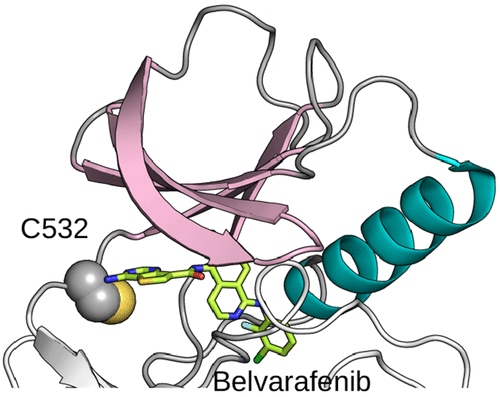
Romany et al, J Chem Inf Model, 2023
We identified druggable cysteines using a novel protocol that combines CpHMD and structural analysis.

-
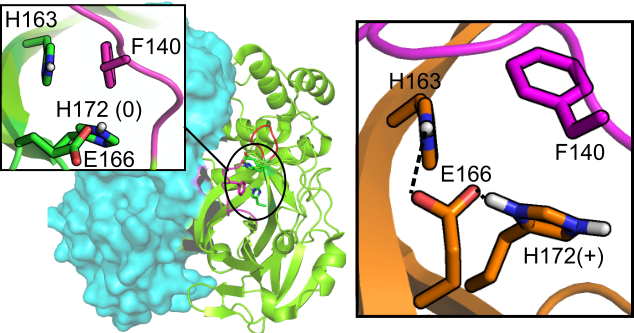
Why is SARS-CoV-2 H172Y Mpro resistant to Paxlovid?
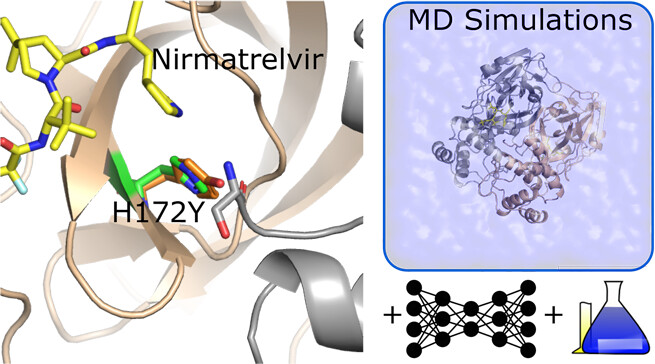
Clayton & Oliverira et al, JCIM, 2023
The drug resistance mechanism of a SARS-CoV-2 Mpro mutant revealed using simulations, AI, & experiments.


-
The GPU all-atom CpHMD is in Amber!
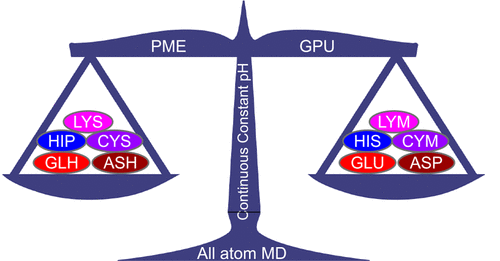
Julie Harris & Ruibin Liu, JCTC, 2022
We present the first CUDA implementation of the all-atom PME CpHMD method in Amber22.


-
Profiling MAPK cysteine reactivity in silico
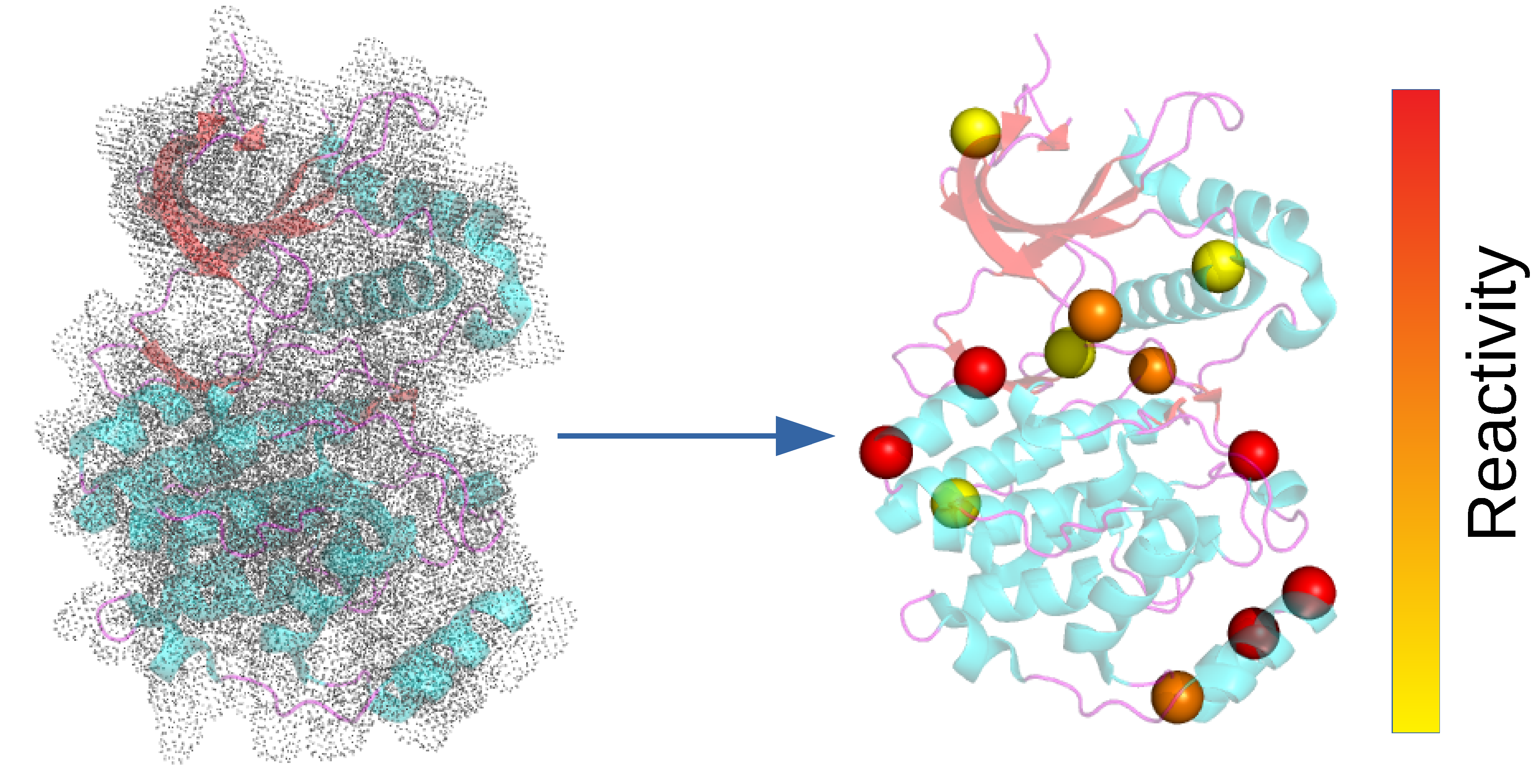
Liu et al, RSC Med Chem, 2021
We assessed and rationalized the cysteine reactivities for all 14 MAP kinases.

-
Kinase front pocket cysteine has various reactivities
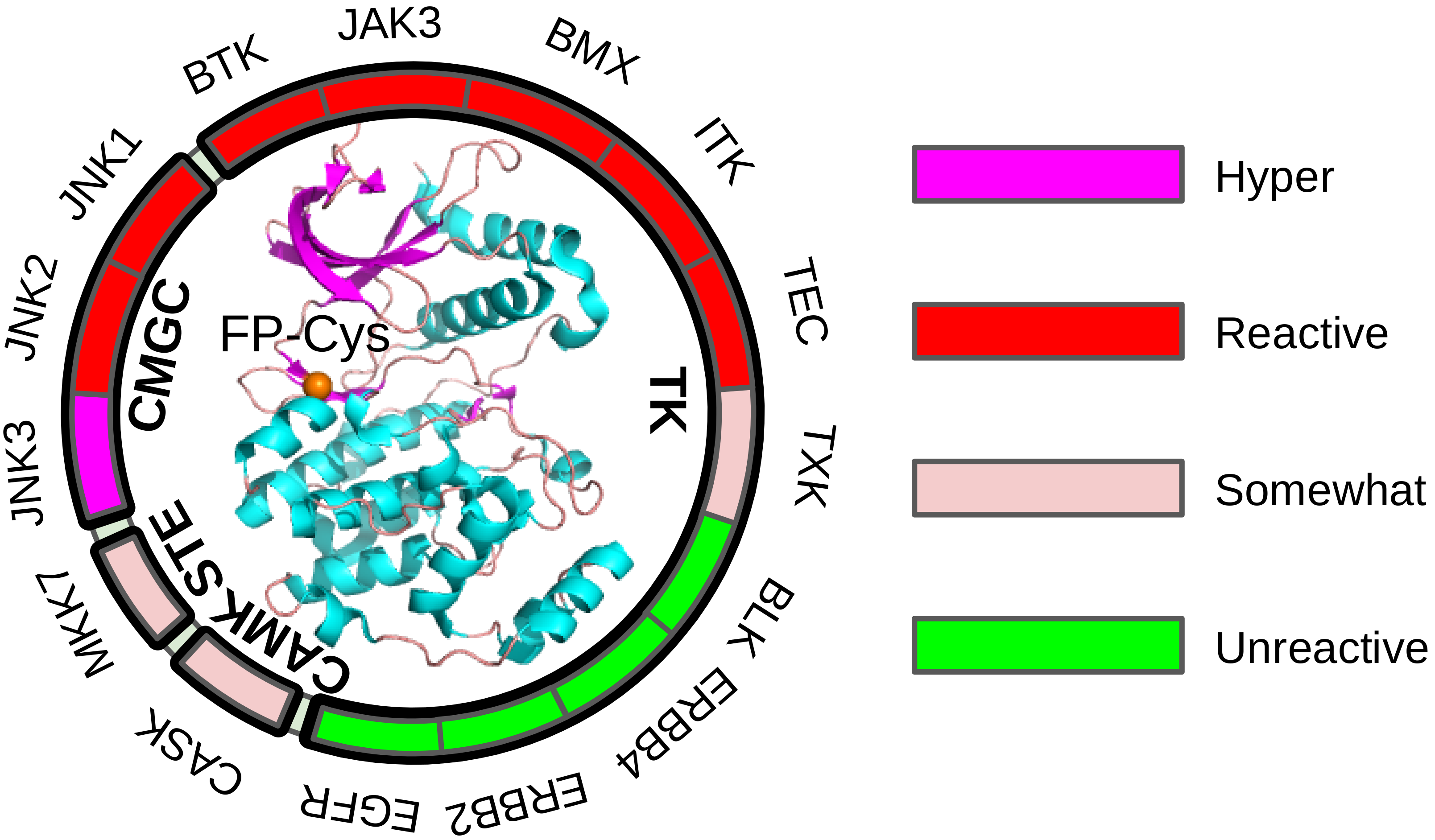
Liu & Zhan et al, J Med Chem, 2021
Exploring the reactivities of the front pocket cysteine in human kinases for covalent drug design.


-
How fentanyl unbinds from mu-opioid receptor
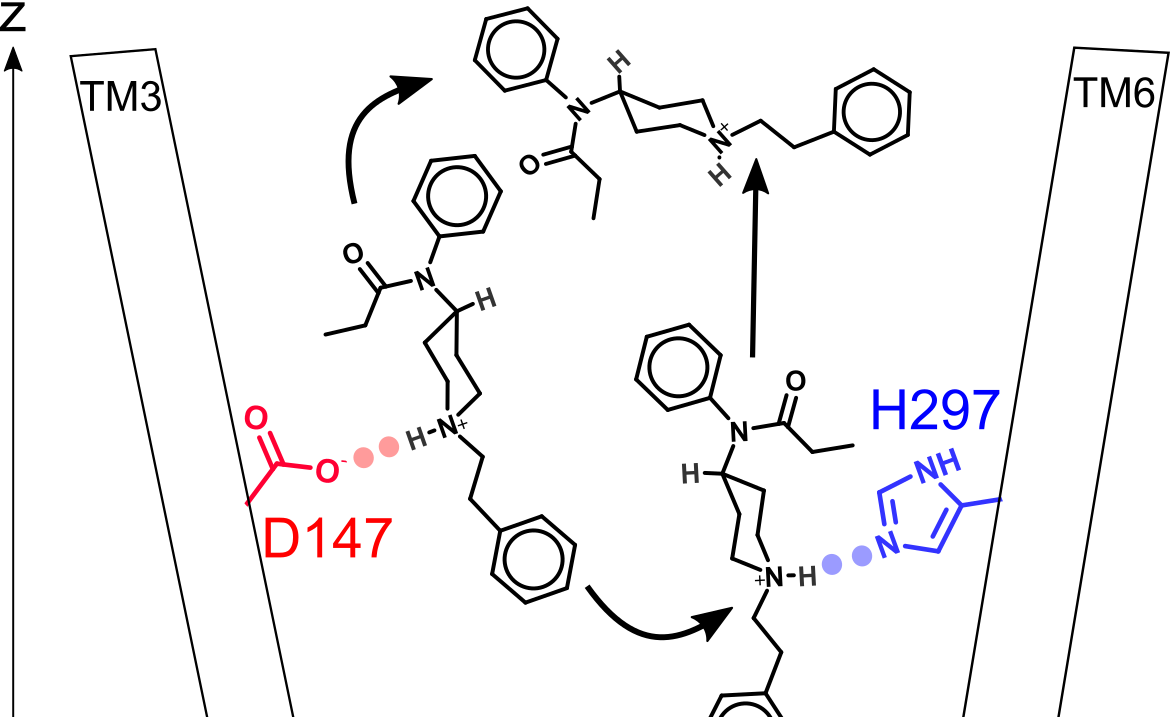
Mahinthichaichan et al, JACS Au 2021
We elucidated the mechanism and kinetics of fentanyl-mOR dissociation to help evaluate drug overdose reversal strategies.

-
Our first effort to combat the opioid crisis!
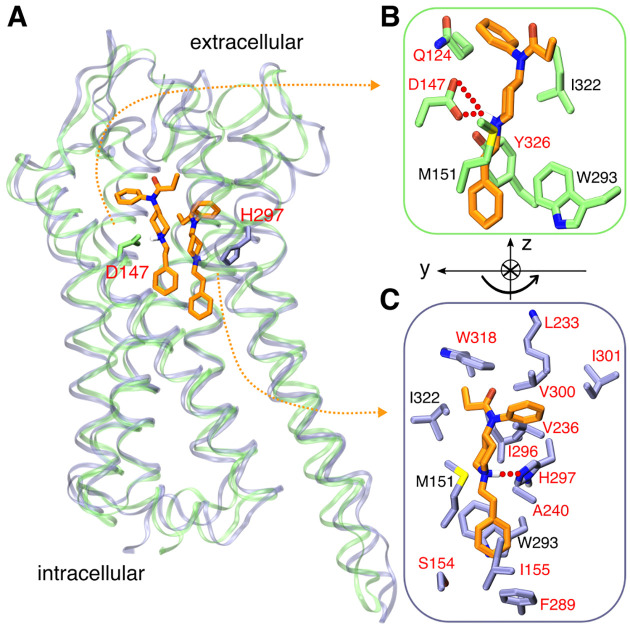
Vo et al, Nat Commun 2021
Deciphering how fentanyl binds mu-opioid receptor represents the first step in understanding the cascade of molecular events leading to addiction.

-
Towards orally available broad-spectrum antiviral drugs
Verma et al, J Am Chem Soc 2020.
Study of SARS main proteases revealed proton-coupled mechanism and an opportunity for small-molecule broad-spectrum antiviral drug design

-
Address the challenge of COVID-19 and beyond
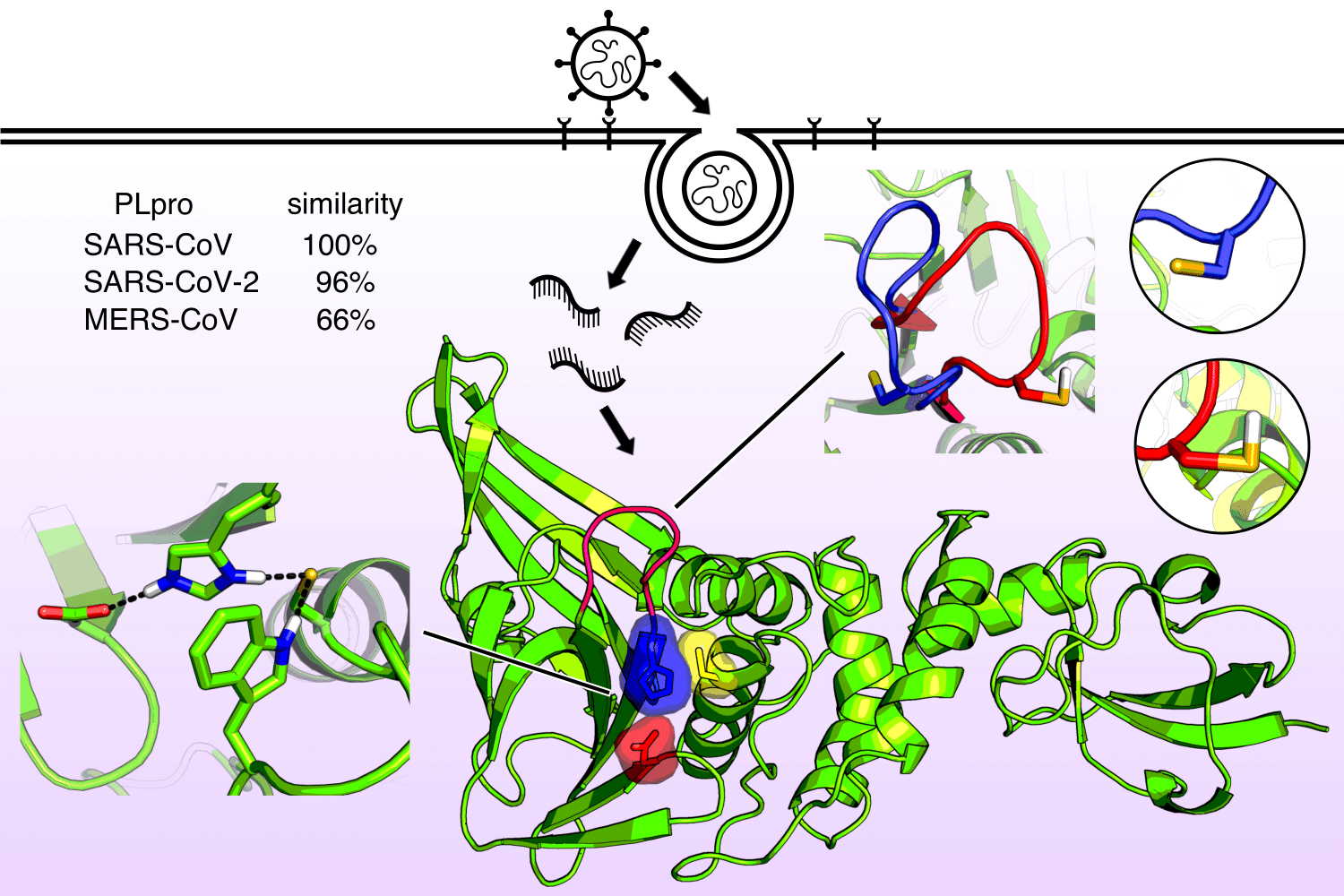
Henderson et al, J Chem Phys, 2020
Studying proton-coupled conformational dynamics of a critical enzyme in SARS and MERS coronaviruses for broad-spectrum antiviral drug design.

-
Nature’s backup plan for ion transporters
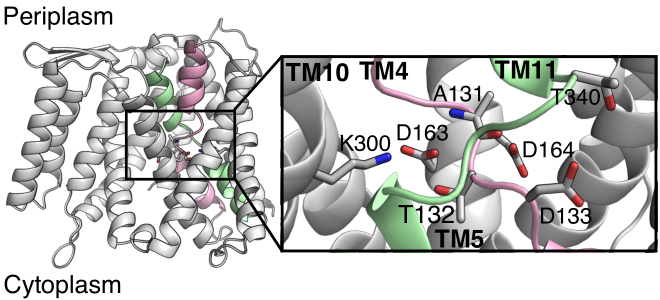
Henderson et al, Proc Natl Acad Sci 2020.
Alternative binding site and proton sharing may represent important general mechanisms of proton-coupled transport in secondary active transporters

-
Accurate prediction of reactive cysteines is possible
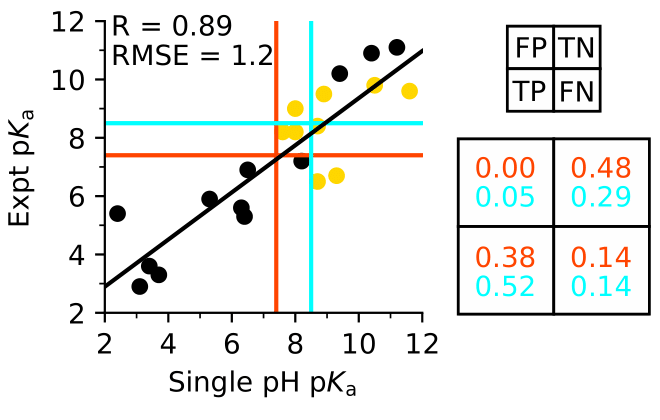
Harris et al, J Chem Theory Comput 2020.
Benchmarking our GPU-based CpHMD tool to facilitate targeted covalent inhibitor design

-
How does an electric field control soft matter?
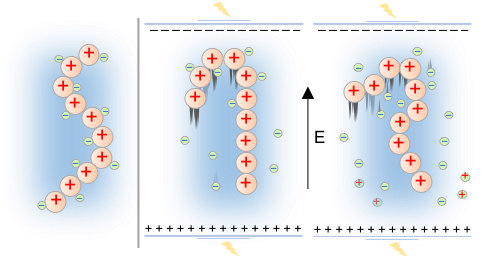
Mahinthichaichan et al, ACS Omega 2020.
Discover how electric signals induce conformational changes of an aminopolysaccharide chain in an unexpected way

-
Persistence. Dedication. A true champion. Congrats to Quynh Vo for completing the JPG puzzle challenge at #bps20 pic.twitter.com/MfFlnsQ2aA
— JGenPhysiol (@JGenPhysiol) February 18, 2020 -
Electrostatic coupling enables conformational plasticity of kinase
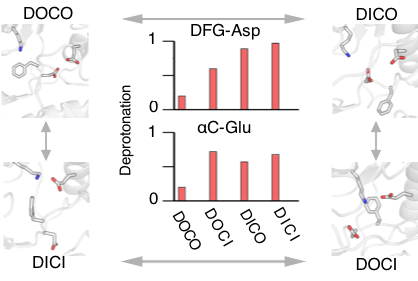
Tsai et al, J Am Chem Soc 2019.
Allowing protonation states to change may be a key to sample the various conformational states of kinases.
-
Accelerating targeted covalent kinase inhibitor design
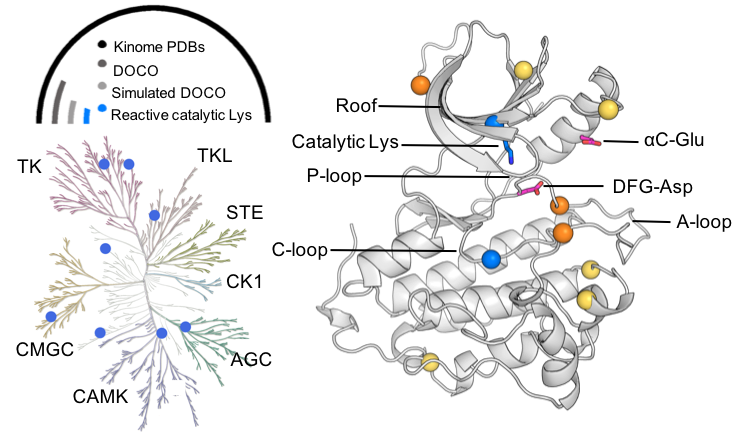
Liu et al, J Am Chem Soc 2019.
A molecular dynamics tool put to test for discovering covalent hotspots in kinases and other drug targets.

-
Continuous constant pH molecular dynamics released in Amber

Harris et al, J Chem Inf Model 2019.
A new GPU implementation for rapid and accurate pKa predictions and proton-coupled dynamics.

-
Modern materials: responsive, reconfigurable, multi-functional
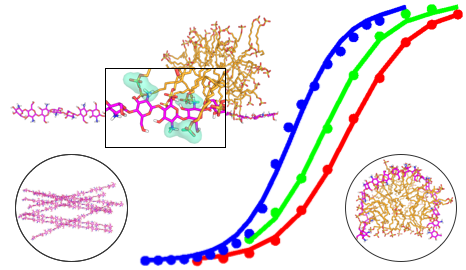
Tsai et al, Chem Mater 2018.
Exploiting a pKa gradient for persistent but erasable gradient in structural and mechanical properties.

-
Ligand protonation state controls water in protein–ligand binding
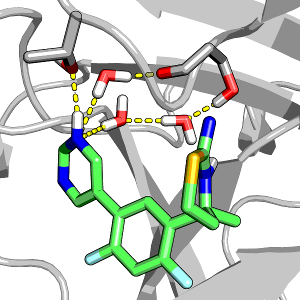
Henderson et al, J Phys Chem Lett 2018.
Pyrimidine protonation shifts the ligand binding mode from water-mediated to direct hydrogen bonding.
